Embryo Report
Advanced genetic screen for your embryos. Prevent your child from inheriting a predisposition to a condition that runs in your family.
Couple Report
Our preconception test measures your future child's genetic predisposition to disease. Mitigate your risk.
For Patients
Our publications
Poster
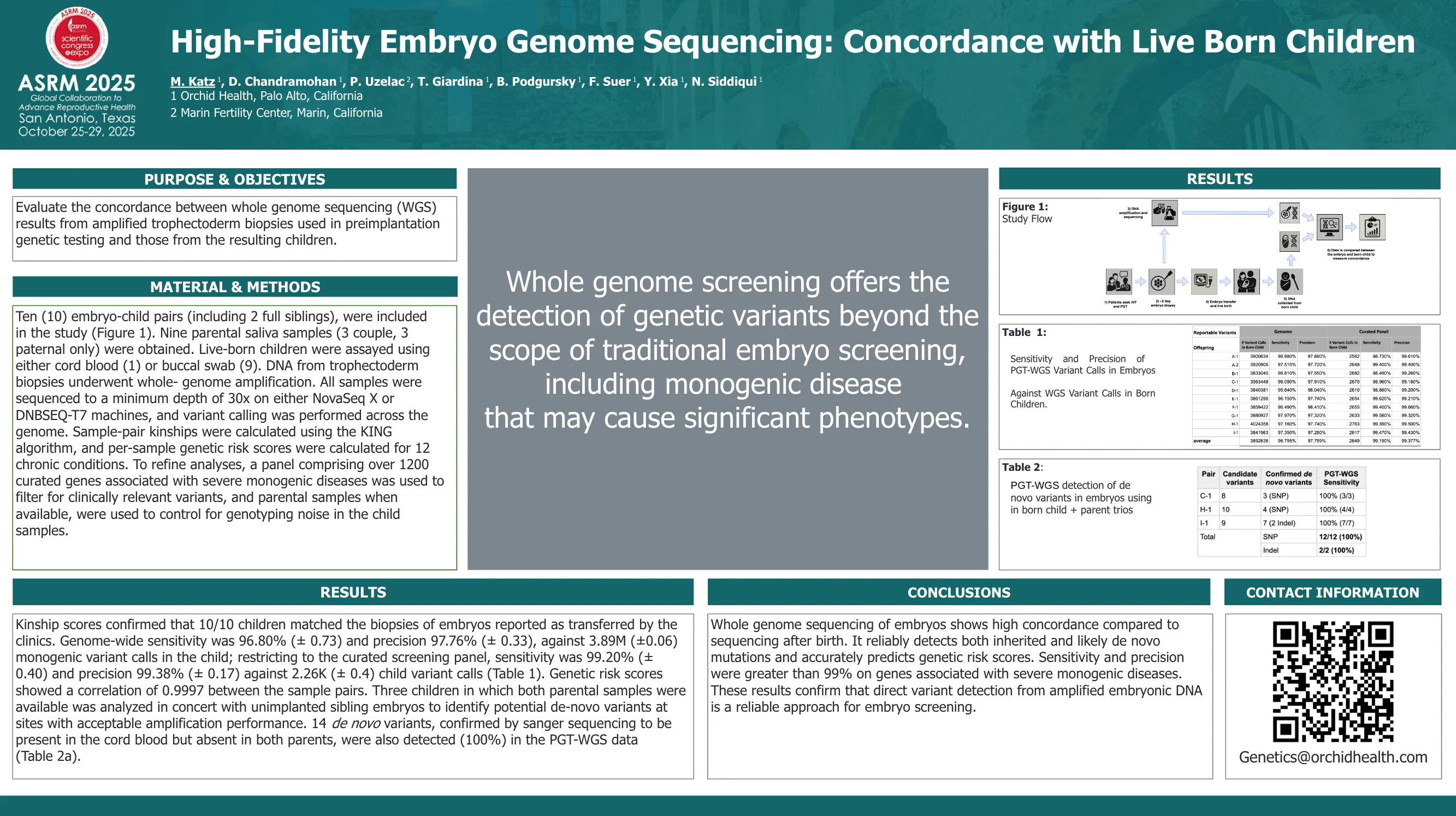
High-Fidelity Embryo Genome Sequencing: Concordance with Live Born Children
Kinship scores confirmed that 10/10 children matched the biopsies of embryos reported as transferred by the clinics. Genome-wide sensitivity was 96.80% (± 0.73) and precision 97.76% (± 0.33), against 3.89M (±0.06) monogenic variant calls in the child...
Presented at: ASRM 2025

Paper
Expanding the horizons of embryo screening: performance and case studies of preimplantation genetic testing via whole genome sequencing
In 2024, we introduced the first PGT—whole genome sequencing (PGT-WGS), enabling the screening of thousands of genes as well as the detection of critical microdeletions and microduplications in one assay. Here, we present assay performance and two cl...
Published: Academia Molecular Biology and Genomics

Poster
Chromosomal Conditions Resolved With Whole Genome Sequencing (WGS) of Embryos
As preimplantation genetic testing (PGT) technologies advance, so too does their resolution and ability to identify conditions that were previously undetectable. High-depth whole genome sequencing (WGS) now provides the capability to screen for chrom...
Presented At: PCRS 2025

Paper
Direct assessment of hereditary hemochromatosis in preimplantation genetic testing
Demonstrating a direct detection and cost-effective approach to detecting HFE C282Y using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) in preimplantation genetic testing (PGT) settings.
Published: F&S Science
Paper
Patient perspectives after receiving simulated preconception polygenic risk scores (PRS) for family planning
The study investigates patient perspectives on the use of Preimplantation Genetic Testing for Polygenic disease (PGT-P) to select embryos with lower risks for common polygenic diseases. Participant responses and attitudes were evaluated after receivi...
Published: Journal of Assisted Reproduction and Genetics

Poster
Preimplantation Genetic Testing (PGT) For De Novo Variants: Embryo Whole Genome Sequencing goes Beyond Carrier Screening Risks
Demonstrating high concordance in exome calls and accurately detecting de novo variants and variants linked to autosomal recessive conditions without the use of probes, PGT-WGS emerges as an invaluable tool for prospective parents
Presented at: NSGC 2024
Preprint
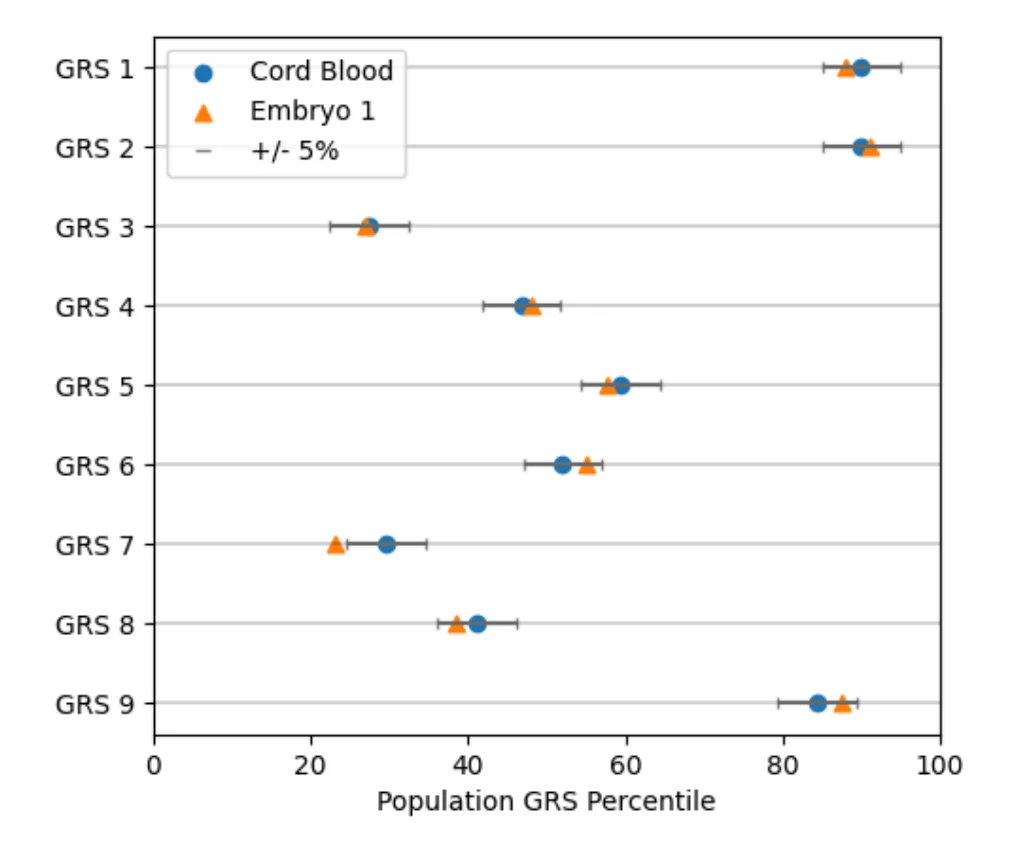
Concordance of whole-genome amplified embryonic DNA with the subsequently born child
Before implantation subsequent to in vitro fertilization (IVF), the current options for Preimplantation Genetic Testing (PGT) are PGT for Aneuploidy (PGT-A) and, if clinically indicated, PGT for monogenic conditions (PGT-M). A more comprehensive appr...
Preprint
Poster
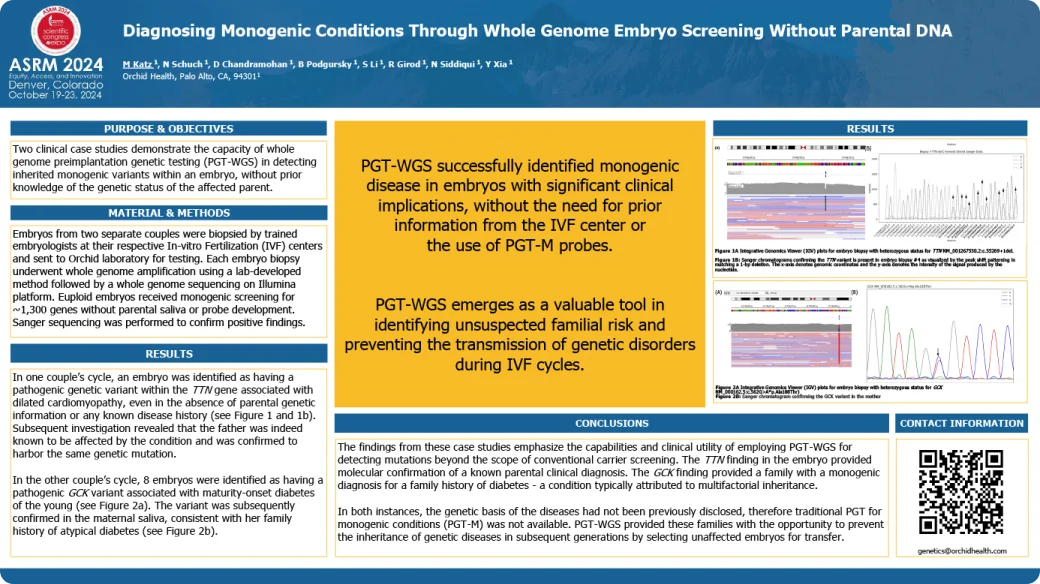
Diagnosing Monogenic Conditions Through Whole Genome Embryo Screening Without Parental DNA
Whole genome embryo screening provides a solution for mitigating the risk of monogenic diseases among patients facing challenging scenarios where traditional PGT for monogenic conditions falls short
Presented at: ASRM 2024

Poster
Challenging PGT-M Cases Resolved with Whole Genome Sequencing (WGS) Of Embryos
PGT-WGS successfully identified monogenic disease in embryos with significant clinical implications, without the need for prior information from the IVF center or the use of PGT-M probes. PGT-WGS emerges as a valuable tool in identifying unsuspected...
Presented at: ASRM 2024
Have healthy babies.
© 2026 Orchid
